The Mullins laboratory pursues answers to fundamental questions in cell biology. In general we ask how nanometer-sized biological molecules transmit and integrate information across much larger length scales: microns, millimeters, and beyond. In other words, we want to understand how collections of macromolecules work together to establish a common identity: to become living cells. One way for molecules to establish long-range order is by self-assembling into much larger structures, such as membranes, cell walls, and cytoskeletal polymers. Our main projects focus on the assembly of cytoskeletal polymer networks and the roles they play in prokaryotic and eukaryotic cells.
Specifically, we ask: (1) How do cytoskeletal structures impose order on the cytoplasm? The molecules in a living cell must all find their proper places and the cytoskeleton plays many roles in this process—establishing landmarks, setting boundaries, and moving cargo. (2) How are cellular signals converted into three-dimensional structures? A signaling pathway can initiate assembly of a cytoskeletal network but we want to understand the molecular mechanisms that specify its architecture and function. (3) What role do cytoskeletal networks play in determining the form and function of multicellular organisms? Events, such as symmetry breaking and polarization, driven by cytoskeletal processes inside cells, are fundamental to determining the body plan of multi-cellular organisms.
Amoeboid cell migration
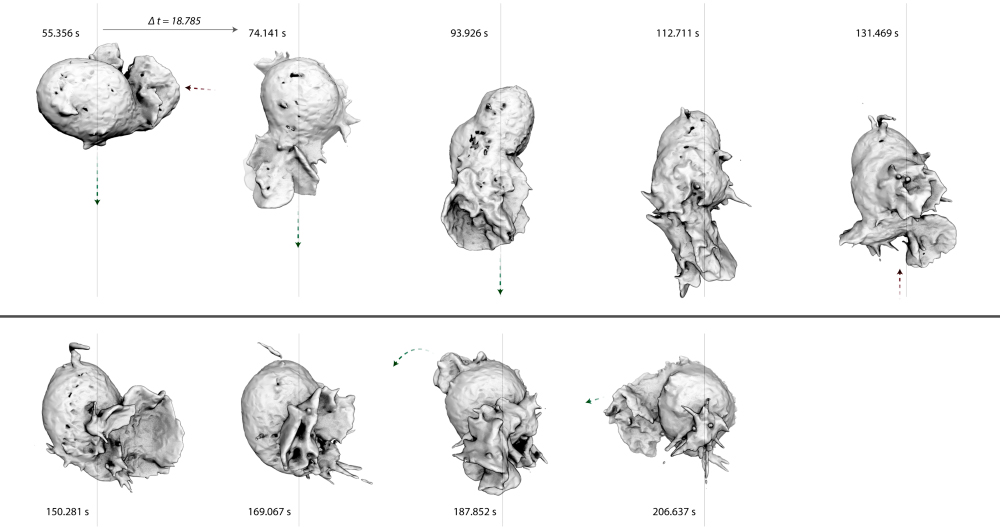
Cell migration is important in single-cell biology, but is also a crucial process in development and cancer metastasis. Despite years of study, the connection between actin filament assembly and amoeboid cell locomotion remains unclear. This, in part, reflects the fact that cell locomotion is not one single process. For years, the canonical view of migration was that of a single sequence of coordinated events: actin-driven membrane protrusion, integrin-mediated leading-edge adhesion, myosin-driven cell body contraction, and force-dependent trailing edge de-adhesion. Recent work, however, has exploded this simple story and we now realize that eukaryotic cells use several different mechanisms to crawl. For instance, some cells (e.g. fast-moving leukocytes) can crawl through complex three-dimensional environments in an integrin-independent manner. The key to this adhesion-independent motility appears to be spatial confinement: when cells are forced to move through restrictions that are small compared to the size of their nuclei, weak electrostatic interactions give them purchase required to move forward. My lab uses advanced imaging techniques to observe crawling dynamics of living cells in order to reveal the complex mechanisms of migration.
Mechano-biochemistry
The mechanical properties of most eukaryotic cells is determined by the actin cytoskeleton. A major challenge to understanding the physical properties of actin networks, however, is that they are dynamic: their assembly and disassembly are integral to their function. At present we know little about how such self-assembling biological materials have evolved to resist and adapt to externally applied forces.
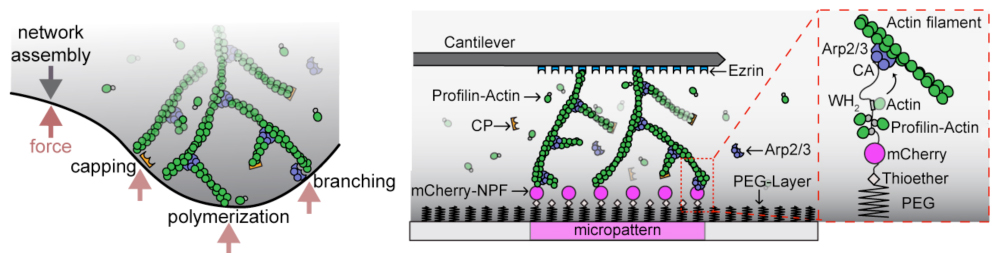
External forces are particularly relevant to ‘dendritic’ actin networks, generated by the nucleating and crosslinking activity of the Arp2/3 complex, a seven-subunit protein complex that builds crosslinked filament arrays by creating new filaments that branch from the sides of existing filaments. Dendritic actin networks assemble in contact with membranes and exert pushing forces that are generated by the free energy of actin polymerization.
To construct force-generating actin networks, the Arp2/3 complex works with a set of highly conserved collaborators: actin, WASP, capping protein, cofilin, and profilin. My lab uses in vitro techniques such as bead-motility assays, AFM, TIRF microscopy, dynamic light scattering, and fluorescence anisotropy to determine the dynamics and biochemistry of these actin networks. We are also are able to apply external forces (using AFM) and observe how actin assembly/disassembly dynamics are perturbed.
Nuclear architecture
Our interest in nuclear actin grew directly out of our work on the Arp2/3 complex, in particular our discovery of a nucleation promoting factor JMY. We found that JMY shuttles into and out of the nucleus in response to DNA damage and cytoplasmic actin assembly.
It has been known since the early 1970s that eukaryotic nuclei contain conventional actin, but the function and form of this nuclear actin have been hotly debated. We are developing new tools and techniques to study nuclear actin. Recently, we have found that DNA damage induces formation of nuclear actin filaments. We are currently studying the assembly and function of these DNA damage-induced nuclear filaments, including whether they alter the mobility of damaged DNA loci or change the rate at which repair factors are recruited to breaks in the DNA.
Bacterial cell biology
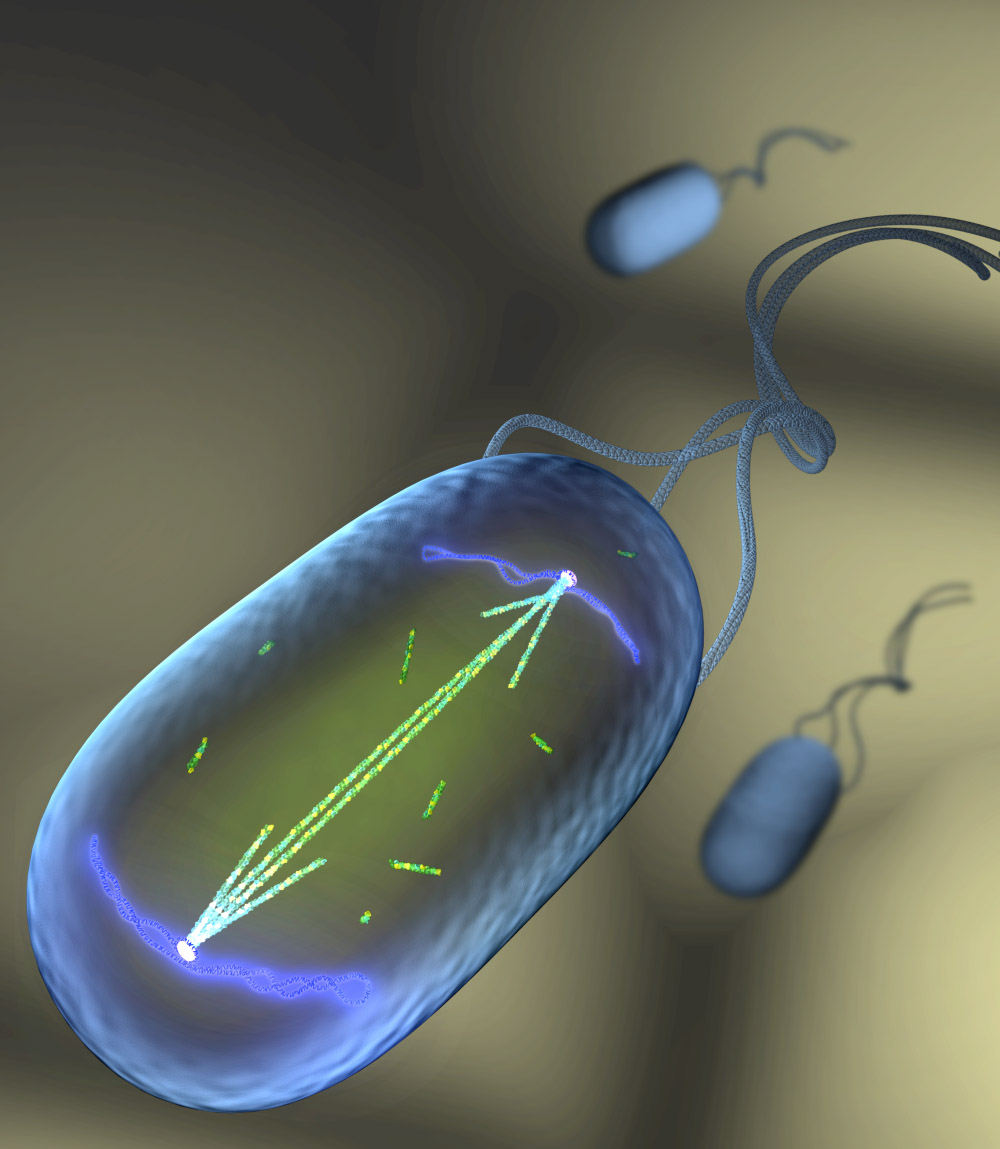
The discovery of cytoskeletal filaments in bacteria defined a new frontier in prokaryotic cell biology. My lab focuses particular attention on movement of intracellular cargo driven by assembly of actin-like ParM filaments. We have characterized the assembly properties of ParM polymers; reconstituted DNA segregation from purified components; and described the dynamics of ParM polymers and cargo DNA molecules in vivo.
We identified three properties of ParM critical for its ability to segregate DNA: fast spontaneous nucleation, symmetrical elongation, and dynamic instability. Of these, the role of dynamic instability is particularly interesting. It underlies three fundamental features of DNA segregation. (1) It ensures that pairs of plasmids are pushed in opposite directions—in effect, counting plasmids. (2) It promotes search and capture by driving plasmids on forced random walks through the cytoplasm, where they encounter other plasmids. (3) And surprisingly, dynamic instability provides energy to push plasmids through the cytoplasm.
We are also working to understand the structure of the interface between ParM filaments and their DNA cargo molecules as well as the mechanism of insertional polymerization that enables cargo complexes to convert the free energy of polymerization into useful work. More recently, we have begun comparing the ParM system with other bacterial, actin-based DNA segregation systems to see whether the rules we have outlined are general or simply one of many solutions to the given problem.
Archaeal cell biology
Archaea represent one of two domains of life on Earth, but we know far less about the cell biology of archaea than we know about bacteria and eukaryotes. For example, we do not understand how most archaea control their shape, organize their intracellular spaces, segregate their DNA, or divide. Mechanistic insight into these processes would shed new light on similar processes in the other domains of life and enable us to better understand the evolution of life on earth and the emergence of the eukaryotic lineage.
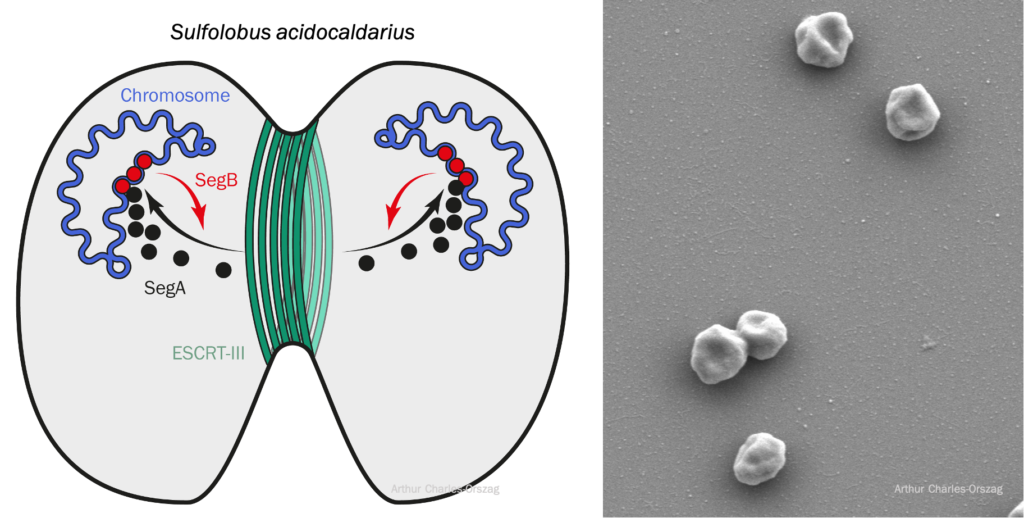
In my lab, we study cell division and DNA segregation in the model crenarchaeon Sulfolobus acidocaldarius that grows optimally at 75–80°C and at pH 2–3. Recently, we developed a system for high-resolution microscopy at high temperature which allowed us to observe live Sulfolobus cells divide at 75°C. We are also working toward reconstituting the conserved SegABs system, which allows chromosome segregation in Sulfolobus. Interestingly, this system resembles the bacterial ParABs system, with SegA being an archaeal homolog of bacterial ParA.